DNA replication in prokaryotes as in the case of E. coli takes at a rate of approximately 1000 nucleotides per second. It means it is a very fast and accurate process of copying the nucleotide bases in metabolically active cells. DNA is synthesized from deoxynucleoside 5'-phosphate precursors (dNTPs such as dGTP, dCTP, dATP and dTTP). It proceeds from 5' end to 3' end. It is because chain growth occurs as a result of the formation of a phosphodiester bond between the 3' oxygen molecule of the growing strand and the alpha-phosphate of a dNTP. An enzyme called DNA polymerase is required to catalyze the reaction. However, a primer is needed to initiate the reaction. Both the strands act as template DNA and the method described here is a semi-conservative method of DNA replication (meaning newly synthesized DNA has one strand from parent DNA). The semi-conservative replication was illustrated by Matthew Meselson and Franklin Stahl in 1958 in an experiment called The Matthew and Stahl experiment. Thus, a primer is required for DNA polymerase to initiate the duplication process. In one strand it is continuous while in the other it is discontinuous. It is because DNA polymerase can only add the elongating DNA strand in 5' to 3'. It is because, with a primer base-paired to the template strand, DNA polymerase adds deoxynucleotides to the free hydroxyl group at the 3' end of the primer as directed by the sequence of the template strand. The place at which a primer anneals with the template DNA is called primer: template junction. Thus, the 3' to 5' end template strand will have a leading strand while the 5' end to 3' end strand has a series of discontinuous fragments of DNA in it called lagging strand and often characterized by Okazaki fragments. Only one primer is needed in the case of the leading strand for its elongation while several primers are needed for the lagging strand till the chain elongation process is accomplished. Prior to the elongation of DNA, these strands need to be separated by the enzyme called helicase which yields a fork called the replication fork. The combination of all proteins that function at the replication fork is referred to as the replisome.
 |
| Fig 1. Molecular Structure of DNA Source: Biorender.com |
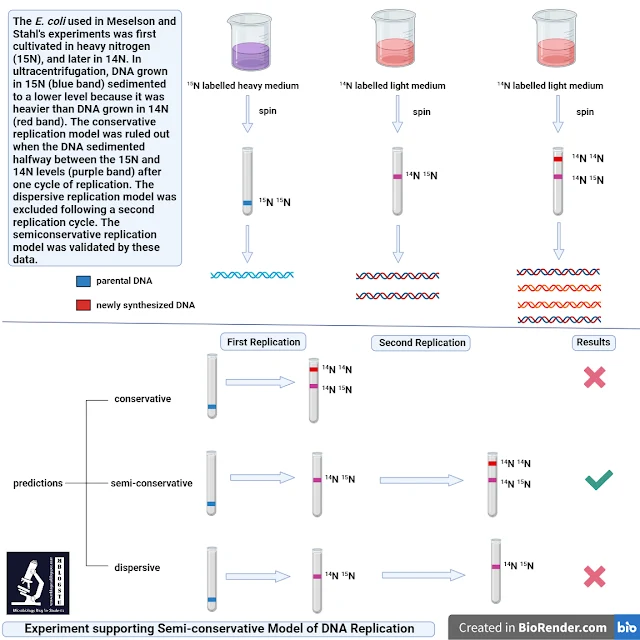
Figure 2. Experiment supporting Semi-conservative model of DNA replication
It is better to describe the process of DNA replication in the following three steps:
1) Initiation
2) Elongation and
3) Termination
Before going on to the aforementioned steps, let's ponder on the essential enzymes required for the DNA replication process in prokaryotes.
i) DNA helicase: It recruits DNA primase to the origin of DNA.
ii) Primase: It helps in synthesizing an RNA primer on each strand of the origin. It also helps in activating the helicase enzyme.
iii) DNA polymerase III holoenzyme: It is brought to the origins through interactions with primer: template junction and the helicase. It helps in catalyzing the elongation of DNA chains during DNA replication.
iv) Single-stranded binding protein (SSB): As a new ssDNA is exposed by the action of the helicases, it is bound by SSBs to avoid DNA rewinding back.
v) Sliding Clamp: Helps to hold the DNA polymerase in place when nucleotides are being added.
vi) Topoisomerases: It helps in maintaining supercoiling in the DNA so that a large number of DNA can be accommodated in a confined region.
vii) DNA polymerase I: Exonuclease activity removes RNA primers and replaces them with newly synthesized DNA.
viii) DNA ligase: Seal the gap between the Okazaki fragments to create one continuous DNA strand.
ix) DNA polymerase II: Repair function
Composition of DNA Pol III holoenzyme
The name holoenzyme suggests a multiprotein complex in which a core enzyme activity is associated with additional components that enhance function. There are four enzymes in each copy of the DNA Pol III holoenzyme. They are three copies of the DNA Pol III core enzyme and one copy of the sliding clamp holder. The sliding clamp holder includes three copies of the Tau protein and each interacts with one DNA Pol III core. There is a presence of a flexible linker between core polymerase and sliding clamp holder/loader and helps in the movement of individual DNA core polymerases during replication.
Proteins involved in E. coli DNA Replication
DnaA (encoded by dnaA)
DnaB (encoded by dnaB)
DnaC (encoded by dnaC)
SSB (encoded by ssb)
Primase (encoded by dnaG)
DNA ligase (encoded by lig)
DNA gyrase
alpha (encoded by gyrA)
beta (encoded by gyrB)
DNA Pol I (encoded by polA)
DNA Pol III (holoenzyme)
alpha (encoded by dnaE)
epsilon (encoded by dnaQ)
theta (encoded by holE)
beta (encoded by dnaN)
Tau a (encoded by dnaX)
gamma b (encoded by dnaX)
delta (encoded by holA)
delta dash (encoded by holB)
chi (encoded by holC)
phi (encoded by holD)
E. coli has 4.6 million base pairs in a single circular chromosome. All of it gets replicated in approximately 42 minutes starting from a single origin of replication and proceeding around the chromosome in both directions. This means that approximately 1000 nucleotides are added per second. Replication in E. coli is explained on the basis of the following points.
1) Initiation
There are specific nucleotide sequences called origins of replication. E. coli has a single origin of replication located on its chromosome. The origin of replication is characterized by approximately 245 base pairs long with AT-rich sequences. This sequence is recognized by certain proteins binding to this site. An enzyme helicase unwinds the DNA by breaking the hydrogen bonds present between the nitrogenous base pairs. This process gets energy by ATP hydrolysis. Following the opening of the DNA helix at the site of origin, two Y-shaped structures are formed called replication forks. Topoisomerase breaks and reforms DNA's phosphate backbone ahead of the replication fork. This helps in relieving the pressure that results from supercoiling. These get extended bi-directionally as replication proceeds. Single-stranded binding proteins prevent each strand of DNA present near the replication fork from winding back into the double helix.
2) Elongation
The next step of elongation where the nucleotides are added sequentially to the growing DNA chain is catalyzed by an important enzyme called DNA polymerase III. The addition of these nucleotides requires energy which is obtained from the nucleotides that have three phosphates attached to them. The energy is released as a result of breaking bonds between the phosphates resulting in the formation of the phosphodiester bond between the incoming nucleotides and the existing chain.
As mentioned earlier, there are three main types of DNA polymerases having specific functions namely DNA polymerase I, DNA polymerase II and DNA polymerase III. DNA polymerase III is required for DNA synthesis. DNA polymerase I is used in the later process of DNA replication while DNA polymerase II is used primarily required for repair.
DNA polymerase is efficient in adding nucleotides only in 5' to 3' direction. However, a free 3'-OH group is required to which the new nucleotide is added by the formation of a phosphodiester bond between the 3'-OH end and the 5' phosphate of the next nucleotide. Thus, another enzyme RNA primase is required for the synthesis of an RNA primer (a 5-10 nucleotide long sequence complementary to the DNA). RNA primase doesn't require a free 3'-OH group. It is because this sequence primes DNA synthesis. After this, the DNA polymerase can now extend this RNA primer adding nucleotides that are complementary to the template strand.
The DNA double helix is anti-parallel in nature meaning that one stand extends from 5' to 3' end while the other from 3' end to 5' end. One strand which is complementary to the 3' end to 5' end parental DNA is synthesized continuously towards the replication fork because the polymerase can add nucleotides in this direction. This continuously synthesized strand is known as the leading strand. The other strand complementary to the 5' end to 3' end parental DNA is extended away from the replication fork in small fragments known as Okazaki fragments. Moreover, Okazaki fragments require a primer to start the synthesis. The strand with the Okazaki fragments is known as the lagging strand. Thus, on the leading strand, DNA is synthesized continuously whereas, on the lagging strand, DNA is synthesized in short stretches called Okazaki fragments. A protein called a sliding clamp holds the DNA polymerase in place during the addition of the nucleotides. The sliding clamp is a ring-shaped protein that binds the DNA and holds the polymerase in place. Topoisomerase prevents the over-winding of the DNA double helix ahead of the replication fork as the DNA is opening up. It does so by causing temporary nicks in the DNA helix and then resealing it. There are two types II topoisomerase in E. coli and they are DNA gyrase and topoisomerase IV (Topo IV). DNA gyrase relaxes positive supercoils ahead of the fork whereas Topo IV decatenates the fully replicated daughter chromosomes. It has also been mentioned that Topo IV helps to resolve pre-catenates and thereby allows fork convergence. DNA polymerase I replaces the RNA primers with DNA while DNA ligase seals the gaps between the Okazaki fragments. This is mediated by joining the fragments into a single DNA molecule. DNA ligase catalyzes the formation of a phosphodiester linkage between the 3'-OH end of one nucleotide and the 5' phosphate end of the other fragment.
3) Termination
The DNA replication is terminated following the convergence of two replication forks. The termination site contains 10 ter sites (A-J). This can bind the DNA replication terminus site-binding protein (Tus) and comprise potent and polar replication fork barriers. However, the function of Tus-ter complexes in replication termination remains unresolved.
References
1. https://openoregon.pressbooks.pub/mhccmajorsbio/chapter/dna-replication-in-prokaryotes/
2. O'Donnell, M., Langston, L., & Stillman, B. (2013). Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harbor perspectives in biology, 5(7), a010108. https://doi.org/10.1101/cshperspect.a010108
3. Dewar, J. M., & Walter, J. C. (2017). Mechanisms of DNA replication termination. Nature reviews. Molecular cell biology, 18(8), 507–516. https://doi.org/10.1038/nrm.2017.42
No comments:
Post a Comment